Abstract
A key challenge in multiple myeloma (MM) is evolution of resistance to standard-of-care (SOC) therapies such as proteasome inhibitors (PI), monoclonal antibodies, and novel targeted therapies. We developed a computational framework that identifies gene programs associated with SOC therapy resistance by pairing patient RNAseq data with clinical response and ex vivo drug response to find patient-specific targets to overcome therapy resistance.
We hypothesize that there are two types of mechanisms of resistance in MM; dynamic (involving cell cycle) and cytostatic (cell-intrinsic, EMDR, complement, etc.). We inferred the dynamic mechanism of resistance by comparing transcriptomic profiles of therapy-naïve vs refractory MM patients, therapy-resistant vs wildtype cell lines, drug perturbation transcriptomics from LINCS-L1000 and activity-based proteomic profiling of MM cell lines. The cytostatic mechanism of resistance was inferred by associating RNAseq data with ex vivo drug response, where CD138+ cells from MM patient samples were co-cultured with human bone marrow stromal cells and patient plasma in an ex vivo reconstruction of the tumor microenvironment and tested with SOC drugs in 399 MM patient samples.
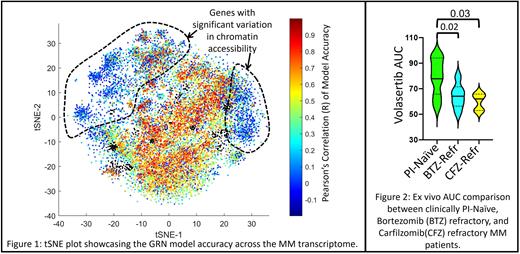
The computational framework begins with the construction of a topological map of MM transcriptome using t-SNE and fuzzy c-means clustering on z-normalized expression of 16,738 genes across 844 MM patient samples, which yielded MM-specific gene programs (clusters of co-expressing genes). Using publicly available ChIP-seq databases like ENCODE and ChEA, we identified regulatory proteins (RPs) upstream of these gene programs. Kinases phosphorylating these RPs were identified from proteomic databases like PhosphoSitePlus and PhosphoPoint; generating a cascading GRN of MM gene programs, their RPs, and kinases, described by a biophysical model representing transcription, translation, and post-translational effects. The model estimates expression of each gene as a function of gene expression of its upstream RPs and kinases. The model for transcriptional regulation for each gene is trained using RNAseq data of 422 patients and validated against the rest of the cohort (422). Figure 1 is a graphical representation of gene-wise Pearson's correlation of model-predicted and actual gene expression represented by the color of each point (gene) on the tSNE. The set of genes that have poor correlation in Figure 1 (blue) statistically significantly (hypergeometric test; p-value <1e-48 and representation factor = 7.27) coincide with genes that are epigenetically regulated (identified by their high variability of chromatin accessibility across MM patients).
We implement this computational framework to identify gene programs associated with a dynamic mechanism of PI (frontline therapy in MM) resistance, which yielded a GRN comprising of 218 co-expressing genes, 13 upstream RPs, and 45 kinases (17 druggable by kinase inhibitors). Network analysis of this GRN revealed that FOXM1 as a key therapeutic vulnerability (characterized by the highest betweenness centrality - a hub for flow of information) that can be targeted by inhibiting PLK1; a key regulator of cell cycle that phosphorylates FOXM1 that, in turn regulates PLK1 expression. A statistically significant (unpaired t-test) increase in z-normalized expression of PLK1 was observed in MM patients (n = 233) clinically refractory to CFZ (p-value < 0.0001) and BTZ (p-value = 0.0004) compared to PI-naïve. Ex vivo functional validation in a cohort of 61 MM patient samples shows a statistically significant (unpaired t-test) lowering of Volasertib (PLK1 inhibitor) AUC in MM patients that were clinically refractory to CFZ (p-value = 0.03) and BTZ (p-value = 0.02) compared to PI-naïve (Figure 2).
Similarly, we infer cytostatic mechanism of resistance to PIs by pairing ex vivo drug response from 399 MM patients with RNAseq data to identify gene programs that are enriched for PI-resistance and PI-sensitivity using gene set enrichment analysis; which identified PRCK, ROCK, and PLK1 inhibitors as therapeutic targets that can either synergize with PIs or exhibit synthetic lethality. Ongoing studies are focused on identifying novel targets to overcome resistance to DARA.
The proposed computational approach can identify novel therapeutic strategies to overcome SOC therapy resistance using patient-specific targeted therapies.
Disclosures
Hampton:M2GEN: Current Employment. Baz:GSK: Honoraria, Membership on an entity's Board of Directors or advisory committees; BMS: Membership on an entity's Board of Directors or advisory committees, Research Funding; Takeda: Membership on an entity's Board of Directors or advisory committees; genentech: Membership on an entity's Board of Directors or advisory committees; Shattuck labs: Membership on an entity's Board of Directors or advisory committees; celgene: Consultancy, Honoraria; karyopharm: Research Funding; Sanofi: Consultancy, Honoraria; Pfizer: Membership on an entity's Board of Directors or advisory committees; Janssen: Membership on an entity's Board of Directors or advisory committees, Research Funding; Merck: Research Funding. Shain:GSK, Janssen and BMS and speaker's bureau for GSK, BMS, Sanofi, Karyopharm, Takeda, Janssen, Adaptive and Amgen: Other: Advisory Committee; Bristol Myers Squibb (BMS), Janssen, GlaxoSmithKline (GSK), Adaptive, Sanofi, and Takeda, and Amgen: Honoraria; Janssen,BMS: Other: PI of clinical trials; AbbVie, Karyopharm: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal